python draw multiple histogram in 3d
Nautical chart Visualization¶
This section demonstrates visualization through charting. For information on visualization of tabular data delight see the section on Tabular array Visualization.
We use the standard convention for referencing the matplotlib API:
In [1]: import matplotlib.pyplot as plt In [2]: plt . close ( "all" ) We provide the basics in pandas to easily create decent looking plots. Meet the ecosystem section for visualization libraries that go beyond the basics documented here.
Notation
All calls to np.random are seeded with 123456.
Bones plotting: plot ¶
We will demonstrate the nuts, see the cookbook for some advanced strategies.
The plot method on Serial and DataFrame is but a simple wrapper around plt.plot() :
In [three]: ts = pd . Series ( np . random . randn ( one thousand ), index = pd . date_range ( "1/1/2000" , periods = 1000 )) In [4]: ts = ts . cumsum () In [5]: ts . plot (); 
If the alphabetize consists of dates, it calls gcf().autofmt_xdate() to attempt to format the x-centrality nicely as per to a higher place.
On DataFrame, plot() is a convenience to plot all of the columns with labels:
In [vi]: df = pd . DataFrame ( np . random . randn ( 1000 , 4 ), index = ts . index , columns = list ( "ABCD" )) In [7]: df = df . cumsum () In [8]: plt . figure (); In [nine]: df . plot (); 
You tin plot 1 column versus some other using the x and y keywords in plot() :
In [x]: df3 = pd . DataFrame ( np . random . randn ( 1000 , 2 ), columns = [ "B" , "C" ]) . cumsum () In [11]: df3 [ "A" ] = pd . Serial ( list ( range ( len ( df )))) In [12]: df3 . plot ( x = "A" , y = "B" ); 
Note
For more formatting and styling options, see formatting below.
Other plots¶
Plotting methods let for a handful of plot styles other than the default line plot. These methods can be provided as the kind keyword argument to plot() , and include:
-
'bar' or 'barh' for bar plots
-
'hist' for histogram
-
'box' for boxplot
-
'kde' or 'density' for density plots
-
'area' for area plots
-
'besprinkle' for scatter plots
-
'hexbin' for hexagonal bin plots
-
'pie' for pie plots
For example, a bar plot tin can be created the post-obit way:
In [thirteen]: plt . figure (); In [14]: df . iloc [ 5 ] . plot ( kind = "bar" ); 
You lot can also create these other plots using the methods DataFrame.plot.<kind> instead of providing the kind keyword argument. This makes it easier to notice plot methods and the specific arguments they use:
In [xv]: df = pd . DataFrame () In [16]: df . plot .< TAB > # noqa: E225, E999 df.plot.area df.plot.barh df.plot.density df.plot.hist df.plot.line df.plot.scatter df.plot.bar df.plot.box df.plot.hexbin df.plot.kde df.plot.pie In addition to these kind due south, at that place are the DataFrame.hist(), and DataFrame.boxplot() methods, which utilise a split up interface.
Finally, in that location are several plotting functions in pandas.plotting that take a Series or DataFrame as an argument. These include:
-
Scatter Matrix
-
Andrews Curves
-
Parallel Coordinates
-
Lag Plot
-
Autocorrelation Plot
-
Bootstrap Plot
-
RadViz
Plots may as well be adorned with errorbars or tables.
Bar plots¶
For labeled, non-time serial data, you may wish to produce a bar plot:
In [17]: plt . figure (); In [18]: df . iloc [ 5 ] . plot . bar (); In [19]: plt . axhline ( 0 , colour = "thou" ); 
Calling a DataFrame's plot.bar() method produces a multiple bar plot:
In [20]: df2 = pd . DataFrame ( np . random . rand ( 10 , 4 ), columns = [ "a" , "b" , "c" , "d" ]) In [21]: df2 . plot . bar (); 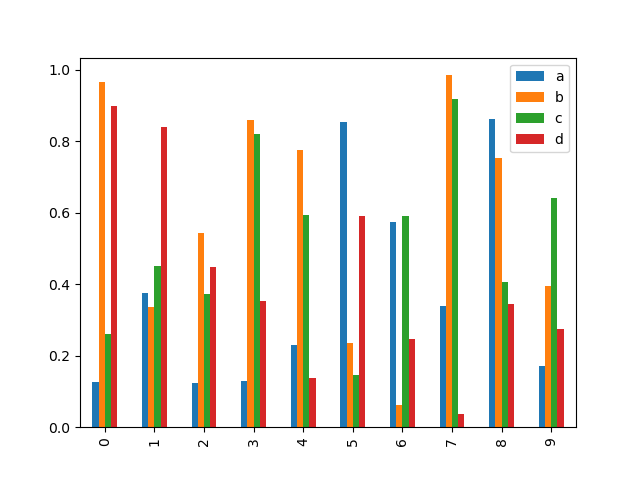
To produce a stacked bar plot, laissez passer stacked=True :
In [22]: df2 . plot . bar ( stacked = Truthful ); 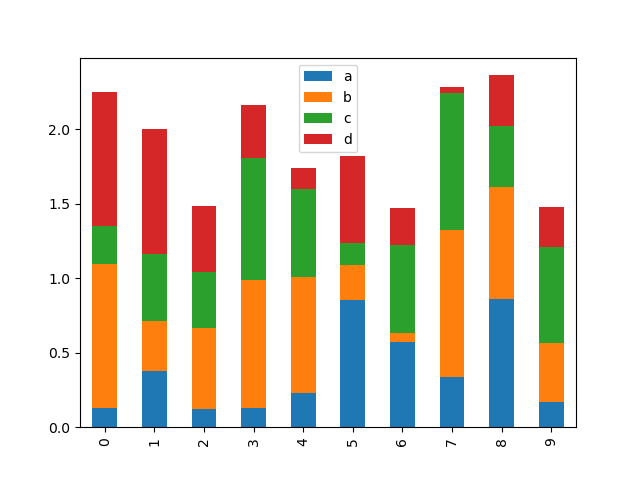
To get horizontal bar plots, use the barh method:
In [23]: df2 . plot . barh ( stacked = True ); 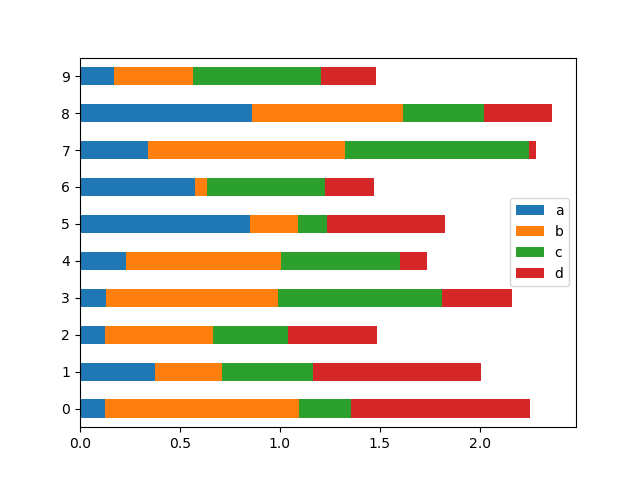
Histograms¶
Histograms tin be fatigued by using the DataFrame.plot.hist() and Serial.plot.hist() methods.
In [24]: df4 = pd . DataFrame ( ....: { ....: "a" : np . random . randn ( 1000 ) + one , ....: "b" : np . random . randn ( 1000 ), ....: "c" : np . random . randn ( 1000 ) - 1 , ....: }, ....: columns = [ "a" , "b" , "c" ], ....: ) ....: In [25]: plt . figure (); In [26]: df4 . plot . hist ( blastoff = 0.5 ); 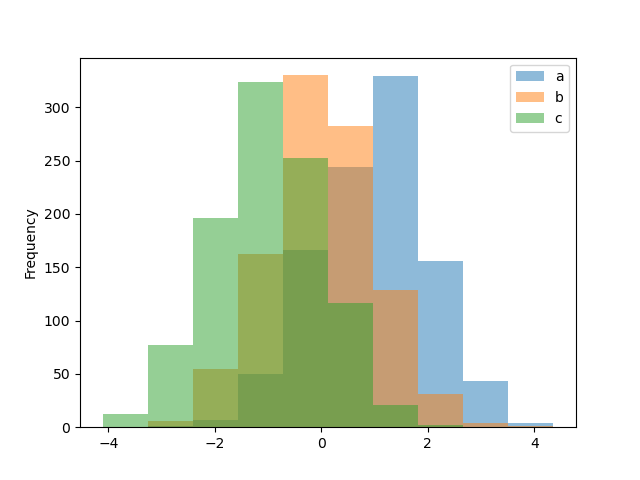
A histogram can be stacked using stacked=True . Bin size tin can be inverse using the bins keyword.
In [27]: plt . effigy (); In [28]: df4 . plot . hist ( stacked = True , bins = 20 ); 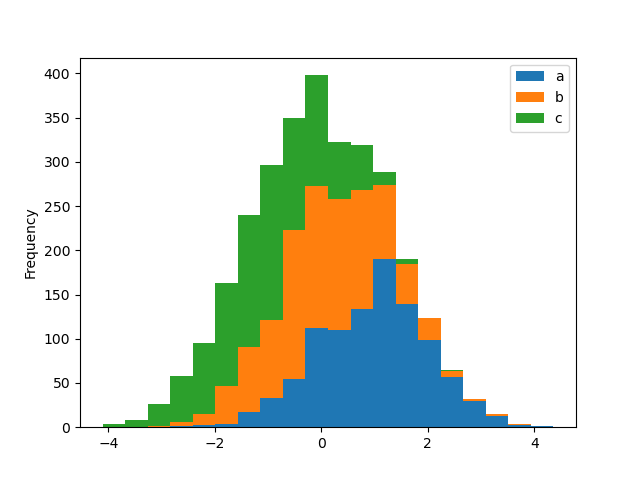
Y'all can laissez passer other keywords supported by matplotlib hist . For example, horizontal and cumulative histograms tin be drawn by orientation='horizontal' and cumulative=True .
In [29]: plt . figure (); In [thirty]: df4 [ "a" ] . plot . hist ( orientation = "horizontal" , cumulative = Truthful ); 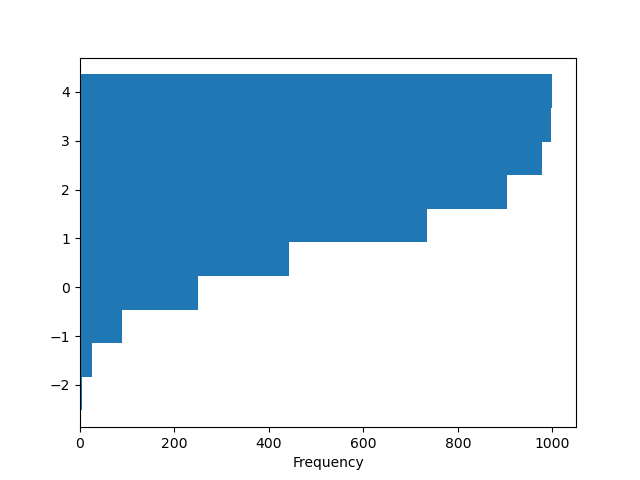
Come across the hist method and the matplotlib hist documentation for more.
The existing interface DataFrame.hist to plot histogram nevertheless can be used.
In [31]: plt . figure (); In [32]: df [ "A" ] . diff () . hist (); 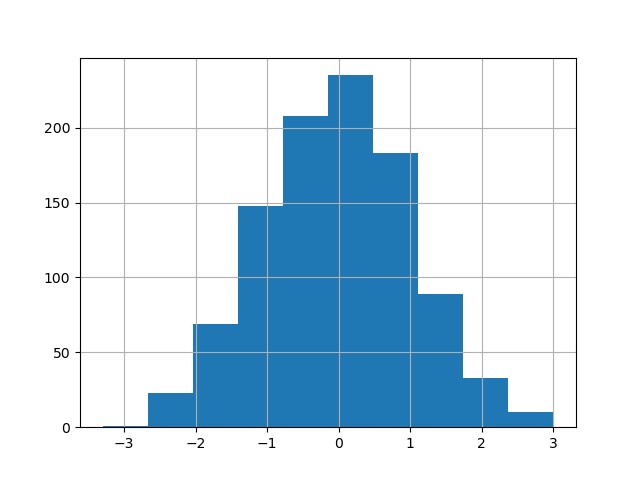
DataFrame.hist() plots the histograms of the columns on multiple subplots:
In [33]: plt . figure (); In [34]: df . diff () . hist ( color = "k" , alpha = 0.5 , bins = 50 ); 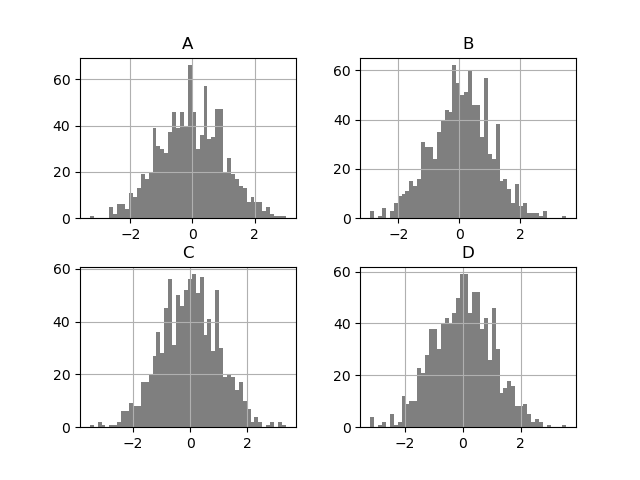
The by keyword can exist specified to plot grouped histograms:
In [35]: information = pd . Series ( np . random . randn ( g )) In [36]: data . hist ( by = np . random . randint ( 0 , 4 , 1000 ), figsize = ( 6 , 4 )); 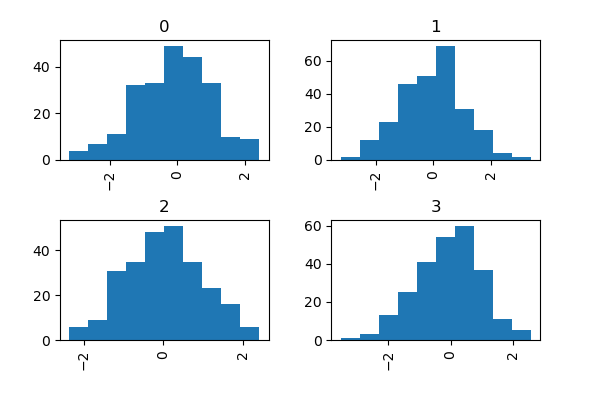
In addition, the by keyword tin can likewise be specified in DataFrame.plot.hist() .
Changed in version ane.4.0.
In [37]: data = pd . DataFrame ( ....: { ....: "a" : np . random . choice ([ "10" , "y" , "z" ], thousand ), ....: "b" : np . random . choice ([ "e" , "f" , "g" ], 1000 ), ....: "c" : np . random . randn ( g ), ....: "d" : np . random . randn ( chiliad ) - 1 , ....: }, ....: ) ....: In [38]: data . plot . hist ( by = [ "a" , "b" ], figsize = ( ten , 5 )); 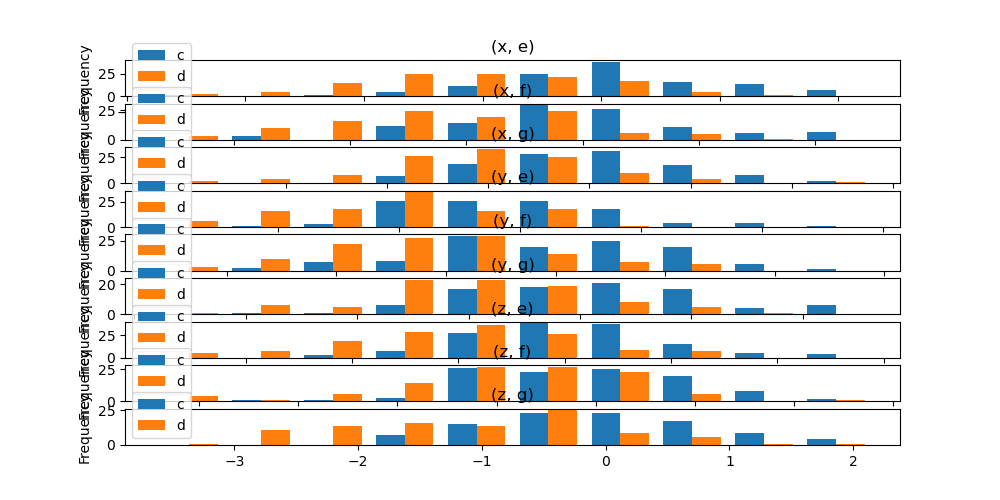
Box plots¶
Boxplot can be fatigued calling Series.plot.box() and DataFrame.plot.box() , or DataFrame.boxplot() to visualize the distribution of values within each cavalcade.
For instance, here is a boxplot representing five trials of 10 observations of a uniform random variable on [0,1).
In [39]: df = pd . DataFrame ( np . random . rand ( 10 , 5 ), columns = [ "A" , "B" , "C" , "D" , "East" ]) In [xl]: df . plot . box (); 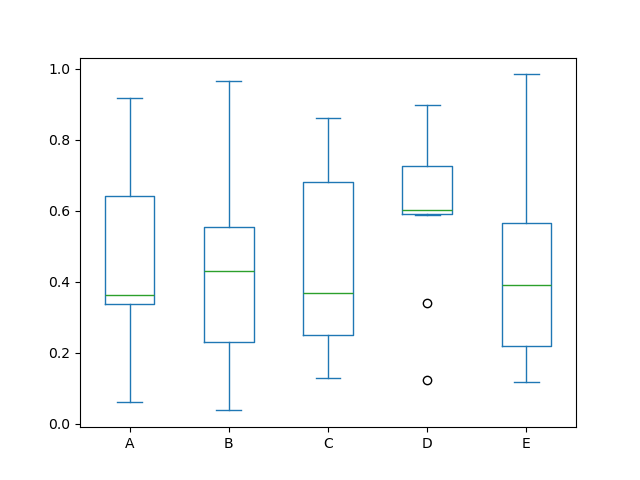
Boxplot can be colorized past passing color keyword. You tin can pass a dict whose keys are boxes , whiskers , medians and caps . If some keys are missing in the dict , default colors are used for the corresponding artists. Likewise, boxplot has sym keyword to specify fliers style.
When you lot pass other type of arguments via colour keyword, information technology will be directly passed to matplotlib for all the boxes , whiskers , medians and caps colorization.
The colors are applied to every boxes to be drawn. If you desire more complicated colorization, you can get each drawn artists by passing return_type.
In [41]: color = { ....: "boxes" : "DarkGreen" , ....: "whiskers" : "DarkOrange" , ....: "medians" : "DarkBlue" , ....: "caps" : "Grey" , ....: } ....: In [42]: df . plot . box ( color = color , sym = "r+" ); 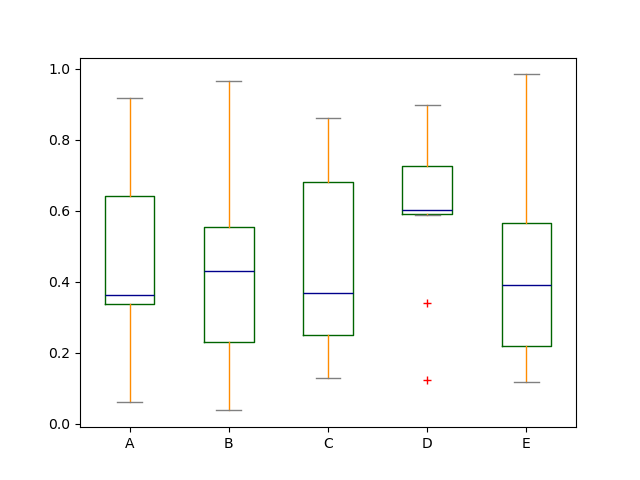
Too, yous can pass other keywords supported past matplotlib boxplot . For example, horizontal and custom-positioned boxplot can be fatigued by vert=Imitation and positions keywords.
In [43]: df . plot . box ( vert = False , positions = [ 1 , 4 , 5 , half dozen , eight ]); 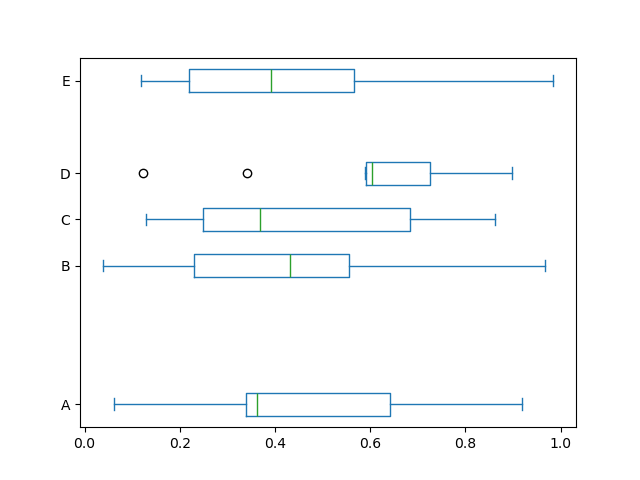
See the boxplot method and the matplotlib boxplot documentation for more than.
The existing interface DataFrame.boxplot to plot boxplot still can be used.
In [44]: df = pd . DataFrame ( np . random . rand ( x , 5 )) In [45]: plt . figure (); In [46]: bp = df . boxplot () 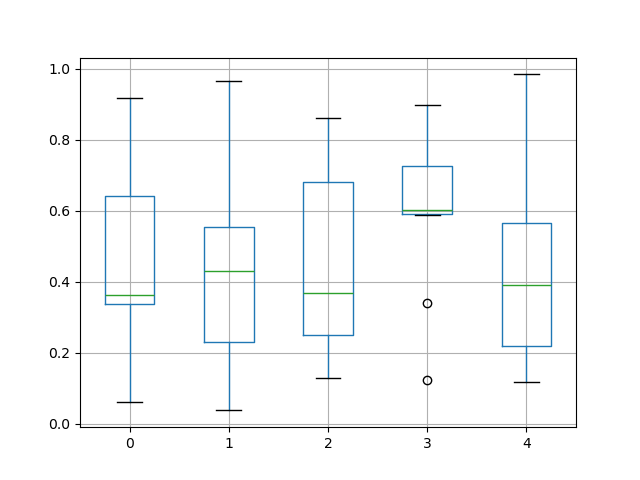
You tin can create a stratified boxplot using the by keyword argument to create groupings. For instance,
In [47]: df = pd . DataFrame ( np . random . rand ( 10 , 2 ), columns = [ "Col1" , "Col2" ]) In [48]: df [ "X" ] = pd . Series ([ "A" , "A" , "A" , "A" , "A" , "B" , "B" , "B" , "B" , "B" ]) In [49]: plt . figure (); In [50]: bp = df . boxplot ( by = "Ten" ) 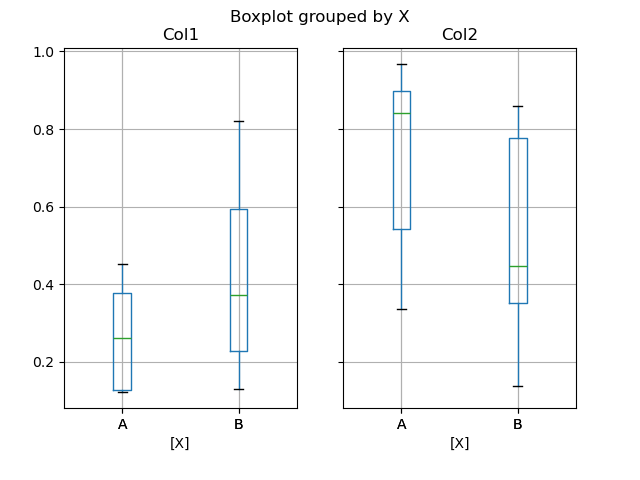
You tin can likewise pass a subset of columns to plot, as well as grouping by multiple columns:
In [51]: df = pd . DataFrame ( np . random . rand ( ten , 3 ), columns = [ "Col1" , "Col2" , "Col3" ]) In [52]: df [ "10" ] = pd . Series ([ "A" , "A" , "A" , "A" , "A" , "B" , "B" , "B" , "B" , "B" ]) In [53]: df [ "Y" ] = pd . Series ([ "A" , "B" , "A" , "B" , "A" , "B" , "A" , "B" , "A" , "B" ]) In [54]: plt . figure (); In [55]: bp = df . boxplot ( cavalcade = [ "Col1" , "Col2" ], past = [ "Ten" , "Y" ]) 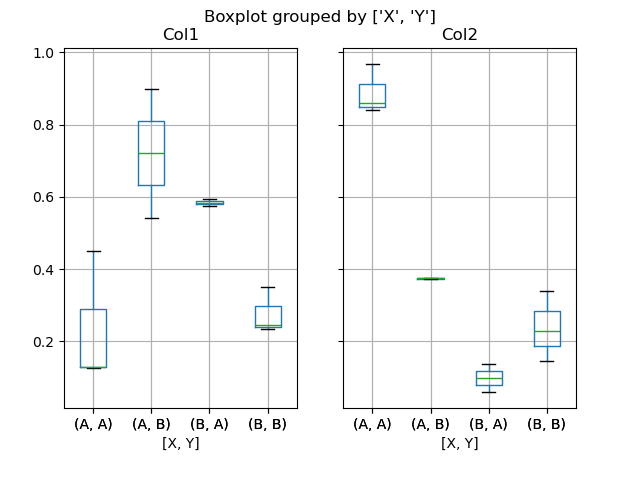
You could besides create groupings with DataFrame.plot.box() , for instance:
Changed in version 1.4.0.
In [56]: df = pd . DataFrame ( np . random . rand ( ten , 3 ), columns = [ "Col1" , "Col2" , "Col3" ]) In [57]: df [ "Ten" ] = pd . Series ([ "A" , "A" , "A" , "A" , "A" , "B" , "B" , "B" , "B" , "B" ]) In [58]: plt . figure (); In [59]: bp = df . plot . box ( column = [ "Col1" , "Col2" ], past = "X" ) 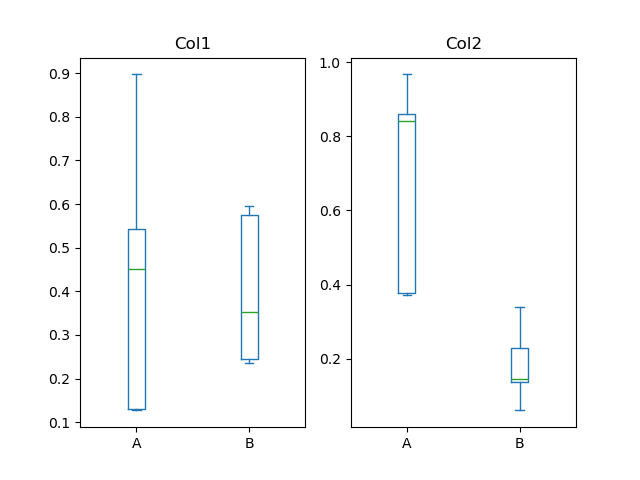
In boxplot , the return blazon tin exist controlled by the return_type , keyword. The valid choices are {"axes", "dict", "both", None} . Faceting, created by DataFrame.boxplot with the by keyword, volition affect the output blazon equally well:
| | Faceted | Output blazon |
|---|---|---|
| | No | axes |
| | Yeah | ii-D ndarray of axes |
| | No | axes |
| | Yep | Series of axes |
| | No | dict of artists |
| | Yes | Series of dicts of artists |
| | No | namedtuple |
| | Yes | Series of namedtuples |
Groupby.boxplot always returns a Serial of return_type .
In [60]: np . random . seed ( 1234 ) In [61]: df_box = pd . DataFrame ( np . random . randn ( 50 , 2 )) In [62]: df_box [ "g" ] = np . random . pick ([ "A" , "B" ], size = 50 ) In [63]: df_box . loc [ df_box [ "g" ] == "B" , 1 ] += iii In [64]: bp = df_box . boxplot ( by = "g" ) 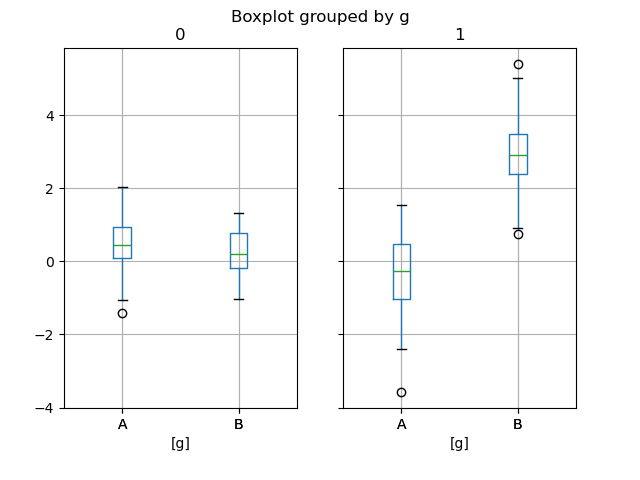
The subplots above are split by the numeric columns first, and so the value of the k cavalcade. Below the subplots are first split past the value of g , then by the numeric columns.
In [65]: bp = df_box . groupby ( "g" ) . boxplot () 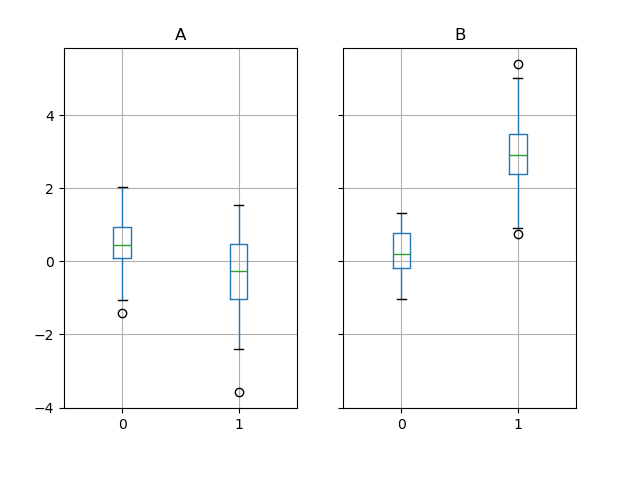
Area plot¶
Yous tin can create expanse plots with Series.plot.area() and DataFrame.plot.area() . Area plots are stacked by default. To produce stacked area plot, each column must be either all positive or all negative values.
When input data contains NaN , it volition be automatically filled by 0. If you want to drop or fill past different values, employ dataframe.dropna() or dataframe.fillna() before calling plot .
In [66]: df = pd . DataFrame ( np . random . rand ( 10 , iv ), columns = [ "a" , "b" , "c" , "d" ]) In [67]: df . plot . expanse (); 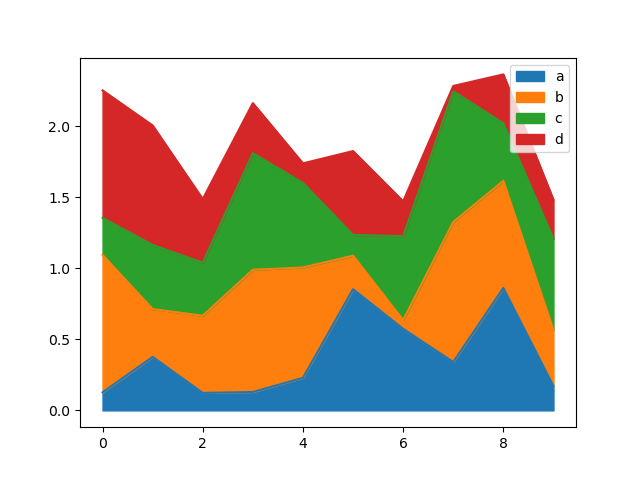
To produce an unstacked plot, laissez passer stacked=False . Alpha value is fix to 0.5 unless otherwise specified:
In [68]: df . plot . area ( stacked = False ); 
Scatter plot¶
Besprinkle plot can be drawn by using the DataFrame.plot.scatter() method. Scatter plot requires numeric columns for the x and y axes. These tin exist specified by the 10 and y keywords.
In [69]: df = pd . DataFrame ( np . random . rand ( l , 4 ), columns = [ "a" , "b" , "c" , "d" ]) In [seventy]: df [ "species" ] = pd . Chiselled ( ....: [ "setosa" ] * xx + [ "versicolor" ] * 20 + [ "virginica" ] * 10 ....: ) ....: In [71]: df . plot . scatter ( x = "a" , y = "b" ); 
To plot multiple column groups in a single axes, repeat plot method specifying target ax . It is recommended to specify color and label keywords to distinguish each groups.
In [72]: ax = df . plot . scatter ( x = "a" , y = "b" , color = "DarkBlue" , label = "Group 1" ) In [73]: df . plot . scatter ( ten = "c" , y = "d" , colour = "DarkGreen" , label = "Group 2" , ax = ax ); 
The keyword c may be given as the name of a column to provide colors for each betoken:
In [74]: df . plot . scatter ( x = "a" , y = "b" , c = "c" , south = 50 ); 
If a chiselled column is passed to c , then a detached colorbar will be produced:
New in version one.3.0.
In [75]: df . plot . scatter ( x = "a" , y = "b" , c = "species" , cmap = "viridis" , s = 50 ); 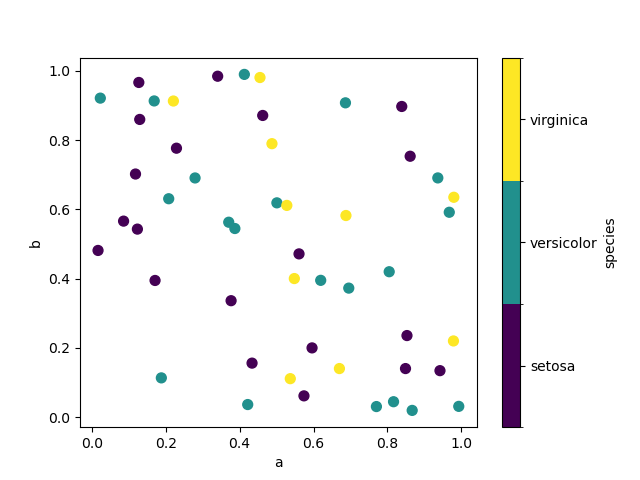
Y'all tin can laissez passer other keywords supported past matplotlib scatter . The example below shows a bubble nautical chart using a column of the DataFrame as the chimera size.
In [76]: df . plot . scatter ( x = "a" , y = "b" , s = df [ "c" ] * 200 ); 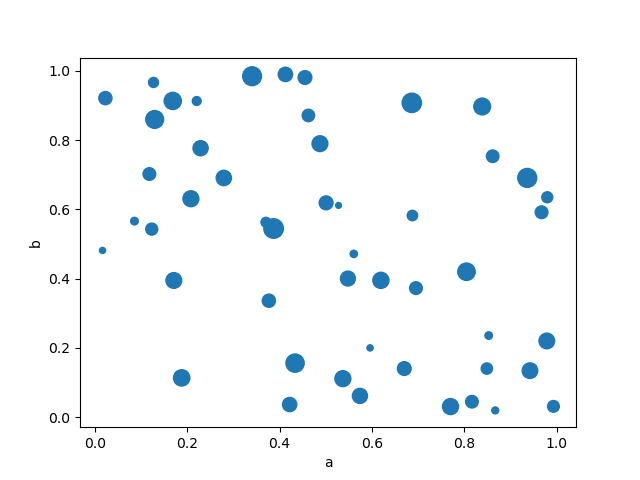
See the besprinkle method and the matplotlib scatter documentation for more.
Hexagonal bin plot¶
Yous can create hexagonal bin plots with DataFrame.plot.hexbin() . Hexbin plots tin be a useful alternative to besprinkle plots if your data are too dense to plot each signal individually.
In [77]: df = pd . DataFrame ( np . random . randn ( 1000 , 2 ), columns = [ "a" , "b" ]) In [78]: df [ "b" ] = df [ "b" ] + np . arange ( 1000 ) In [79]: df . plot . hexbin ( 10 = "a" , y = "b" , gridsize = 25 ); 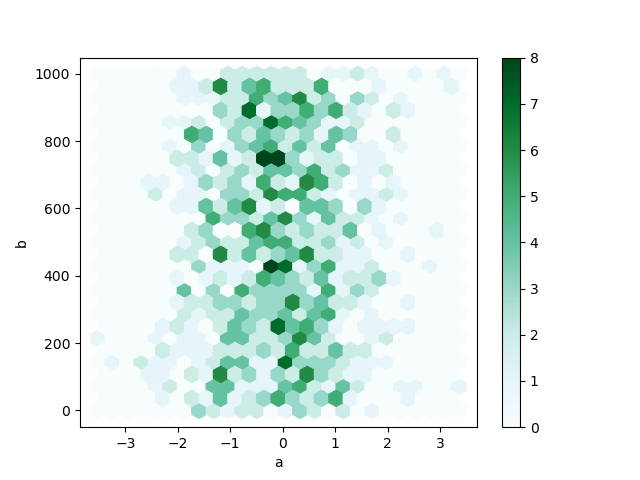
A useful keyword argument is gridsize ; it controls the number of hexagons in the x-management, and defaults to 100. A larger gridsize means more than, smaller bins.
By default, a histogram of the counts around each (10, y) point is computed. You tin specify alternative aggregations by passing values to the C and reduce_C_function arguments. C specifies the value at each (10, y) point and reduce_C_function is a role of one argument that reduces all the values in a bin to a single number (e.g. mean , max , sum , std ). In this case the positions are given by columns a and b , while the value is given by column z . The bins are aggregated with NumPy's max office.
In [80]: df = pd . DataFrame ( np . random . randn ( 1000 , ii ), columns = [ "a" , "b" ]) In [81]: df [ "b" ] = df [ "b" ] + np . arange ( 1000 ) In [82]: df [ "z" ] = np . random . compatible ( 0 , 3 , 1000 ) In [83]: df . plot . hexbin ( x = "a" , y = "b" , C = "z" , reduce_C_function = np . max , gridsize = 25 ); 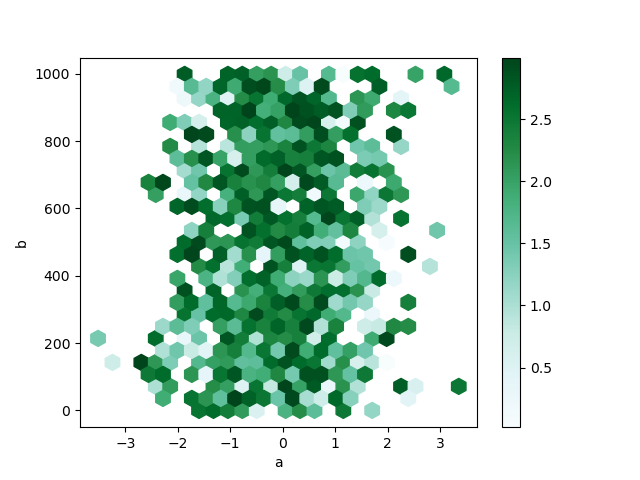
Come across the hexbin method and the matplotlib hexbin documentation for more.
Pie plot¶
Y'all can create a pie plot with DataFrame.plot.pie() or Series.plot.pie() . If your data includes any NaN , they will be automatically filled with 0. A ValueError will exist raised if in that location are any negative values in your data.
In [84]: series = pd . Series ( 3 * np . random . rand ( 4 ), index = [ "a" , "b" , "c" , "d" ], name = "series" ) In [85]: series . plot . pie ( figsize = ( 6 , 6 )); 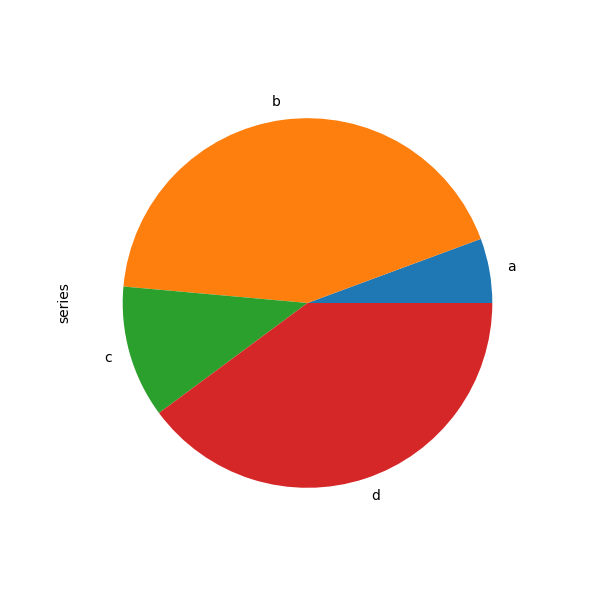
For pie plots it's all-time to use square figures, i.due east. a figure aspect ratio ane. You lot can create the figure with equal width and height, or strength the aspect ratio to be equal afterwards plotting by calling ax.set_aspect('equal') on the returned axes object.
Note that pie plot with DataFrame requires that you either specify a target column by the y argument or subplots=True . When y is specified, pie plot of selected cavalcade volition be drawn. If subplots=Truthful is specified, pie plots for each column are fatigued as subplots. A fable will be drawn in each pie plots by default; specify legend=Fake to hibernate information technology.
In [86]: df = pd . DataFrame ( ....: 3 * np . random . rand ( 4 , 2 ), index = [ "a" , "b" , "c" , "d" ], columns = [ "x" , "y" ] ....: ) ....: In [87]: df . plot . pie ( subplots = True , figsize = ( 8 , 4 )); 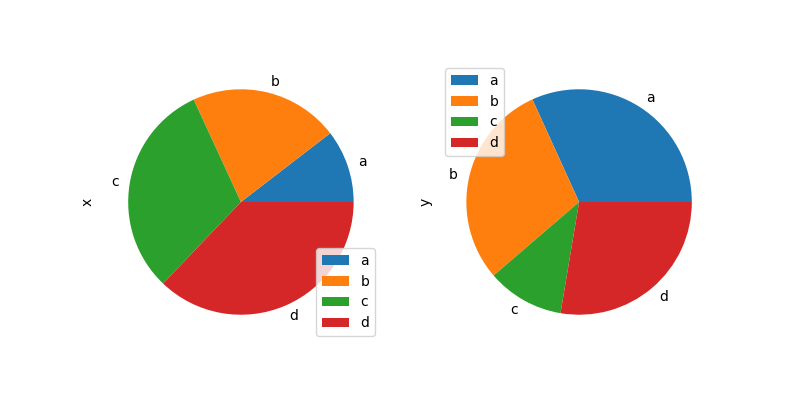
You can employ the labels and colors keywords to specify the labels and colors of each wedge.
Alarm
Almost pandas plots utilise the label and color arguments (note the lack of "s" on those). To be consistent with matplotlib.pyplot.pie() you must utilise labels and colors .
If you want to hide wedge labels, specify labels=None . If fontsize is specified, the value volition exist practical to wedge labels. Also, other keywords supported past matplotlib.pyplot.pie() can exist used.
In [88]: series . plot . pie ( ....: labels = [ "AA" , "BB" , "CC" , "DD" ], ....: colors = [ "r" , "grand" , "b" , "c" ], ....: autopct = " %.2f " , ....: fontsize = 20 , ....: figsize = ( 6 , half-dozen ), ....: ); ....: 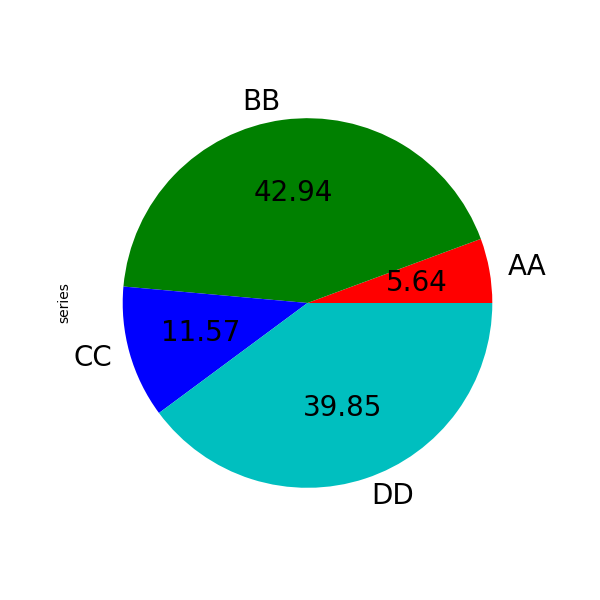
If you laissez passer values whose sum total is less than 1.0, matplotlib draws a semicircle.
In [89]: series = pd . Series ([ 0.i ] * 4 , index = [ "a" , "b" , "c" , "d" ], name = "series2" ) In [90]: serial . plot . pie ( figsize = ( 6 , 6 )); 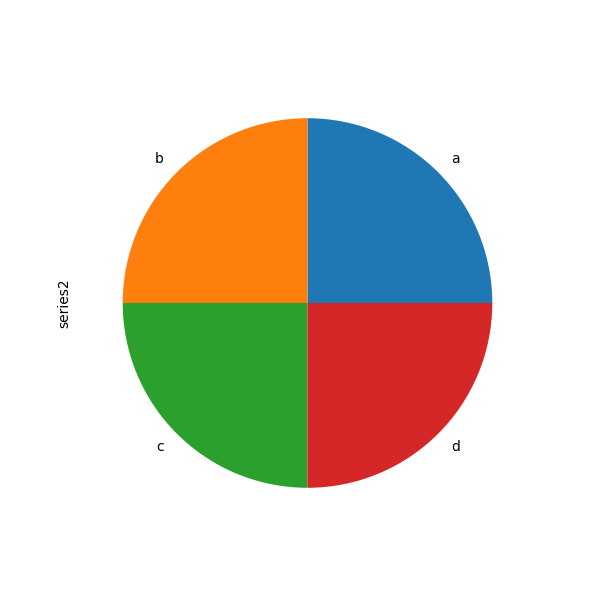
See the matplotlib pie documentation for more.
Plotting with missing information¶
pandas tries to be pragmatic about plotting DataFrames or Serial that contain missing data. Missing values are dropped, left out, or filled depending on the plot type.
| Plot Blazon | NaN Handling |
|---|---|
| Line | Leave gaps at NaNs |
| Line (stacked) | Make full 0'southward |
| Bar | Fill 0's |
| Besprinkle | Drib NaNs |
| Histogram | Driblet NaNs (column-wise) |
| Box | Drop NaNs (cavalcade-wise) |
| Expanse | Fill up 0's |
| KDE | Drop NaNs (cavalcade-wise) |
| Hexbin | Drib NaNs |
| Pie | Fill 0'due south |
If any of these defaults are not what you want, or if you lot desire to be explicit about how missing values are handled, consider using fillna() or dropna() before plotting.
Plotting tools¶
These functions tin be imported from pandas.plotting and have a Series or DataFrame as an argument.
Scatter matrix plot¶
You can create a besprinkle plot matrix using the scatter_matrix method in pandas.plotting :
In [91]: from pandas.plotting import scatter_matrix In [92]: df = pd . DataFrame ( np . random . randn ( m , four ), columns = [ "a" , "b" , "c" , "d" ]) In [93]: scatter_matrix ( df , alpha = 0.2 , figsize = ( half-dozen , vi ), diagonal = "kde" ); 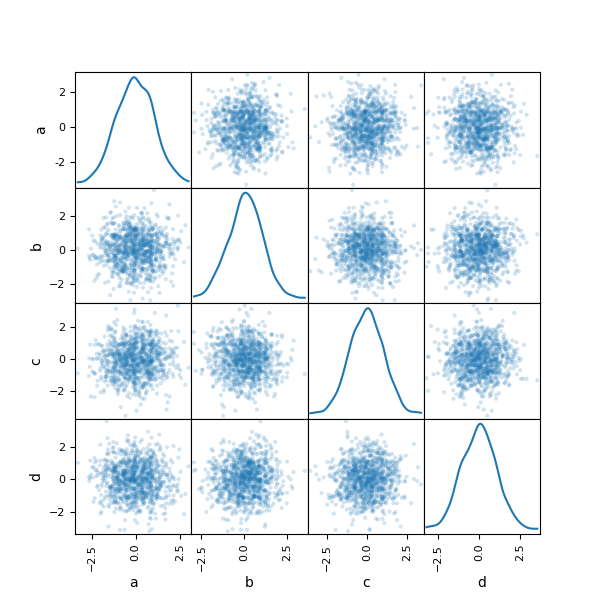
Density plot¶
Y'all tin create density plots using the Series.plot.kde() and DataFrame.plot.kde() methods.
In [94]: ser = pd . Series ( np . random . randn ( m )) In [95]: ser . plot . kde (); 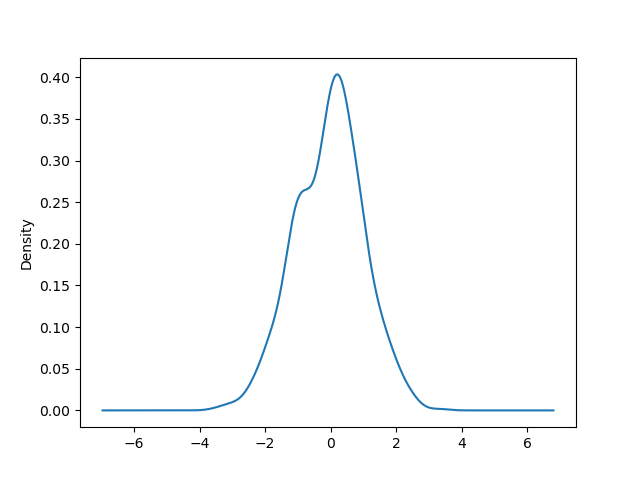
Andrews curves¶
Andrews curves allow one to plot multivariate data as a large number of curves that are created using the attributes of samples as coefficients for Fourier series, come across the Wikipedia entry for more data. Past coloring these curves differently for each class information technology is possible to visualize data clustering. Curves belonging to samples of the same class will usually be closer together and form larger structures.
Note: The "Iris" dataset is bachelor here.
In [96]: from pandas.plotting import andrews_curves In [97]: information = pd . read_csv ( "data/iris.data" ) In [98]: plt . figure (); In [99]: andrews_curves ( information , "Name" ); 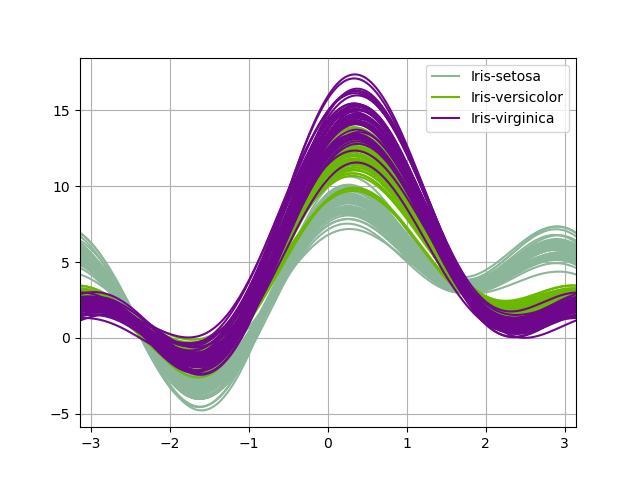
Parallel coordinates¶
Parallel coordinates is a plotting technique for plotting multivariate data, see the Wikipedia entry for an introduction. Parallel coordinates allows i to run across clusters in information and to estimate other statistics visually. Using parallel coordinates points are represented equally continued line segments. Each vertical line represents one aspect. One set of connected line segments represents i data point. Points that tend to cluster volition appear closer together.
In [100]: from pandas.plotting import parallel_coordinates In [101]: data = pd . read_csv ( "data/iris.data" ) In [102]: plt . effigy (); In [103]: parallel_coordinates ( information , "Name" ); 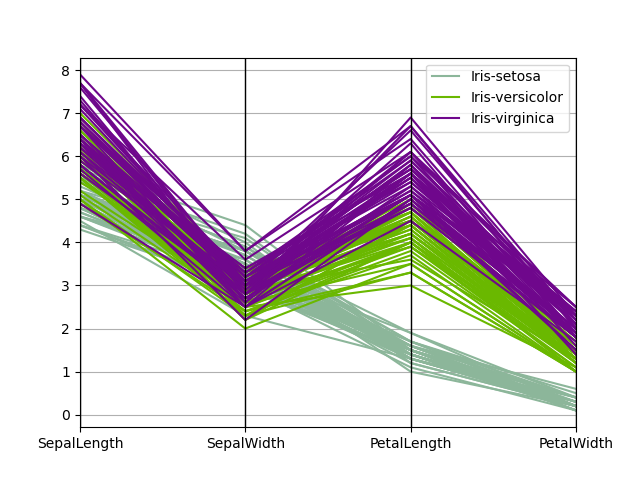
Lag plot¶
Lag plots are used to cheque if a data set or time series is random. Random data should not exhibit any structure in the lag plot. Non-random construction implies that the underlying data are not random. The lag argument may be passed, and when lag=1 the plot is essentially data[:-i] vs. data[1:] .
In [104]: from pandas.plotting import lag_plot In [105]: plt . effigy (); In [106]: spacing = np . linspace ( - 99 * np . pi , 99 * np . pi , num = 1000 ) In [107]: data = pd . Series ( 0.1 * np . random . rand ( thousand ) + 0.9 * np . sin ( spacing )) In [108]: lag_plot ( information ); 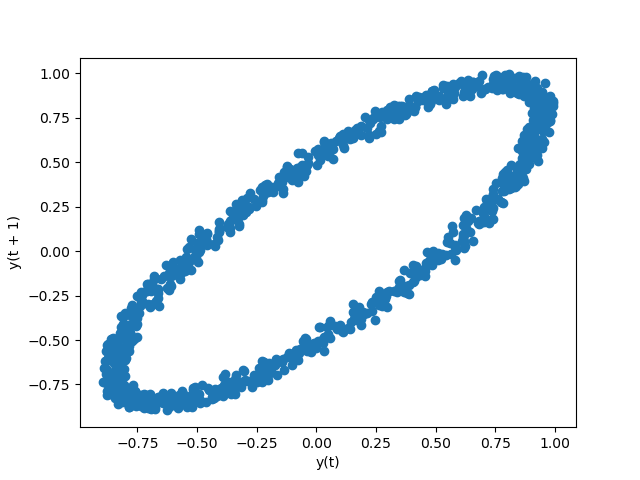
Autocorrelation plot¶
Autocorrelation plots are oftentimes used for checking randomness in fourth dimension series. This is done by computing autocorrelations for data values at varying time lags. If time series is random, such autocorrelations should be about nothing for whatever and all time-lag separations. If time series is non-random then one or more of the autocorrelations will exist significantly non-cypher. The horizontal lines displayed in the plot correspond to 95% and 99% confidence bands. The dashed line is 99% confidence band. See the Wikipedia entry for more nigh autocorrelation plots.
In [109]: from pandas.plotting import autocorrelation_plot In [110]: plt . figure (); In [111]: spacing = np . linspace ( - 9 * np . pi , 9 * np . pi , num = one thousand ) In [112]: data = pd . Serial ( 0.7 * np . random . rand ( 1000 ) + 0.3 * np . sin ( spacing )) In [113]: autocorrelation_plot ( data ); 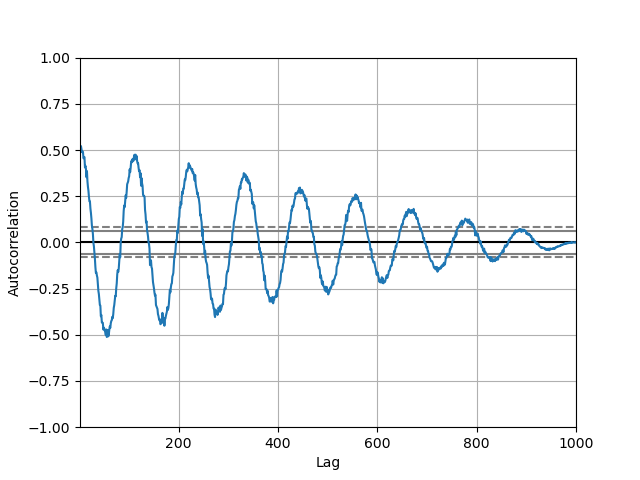
Bootstrap plot¶
Bootstrap plots are used to visually assess the uncertainty of a statistic, such equally mean, median, midrange, etc. A random subset of a specified size is selected from a data set, the statistic in question is computed for this subset and the process is repeated a specified number of times. Resulting plots and histograms are what constitutes the bootstrap plot.
In [114]: from pandas.plotting import bootstrap_plot In [115]: data = pd . Series ( np . random . rand ( thousand )) In [116]: bootstrap_plot ( data , size = l , samples = 500 , color = "grayness" ); 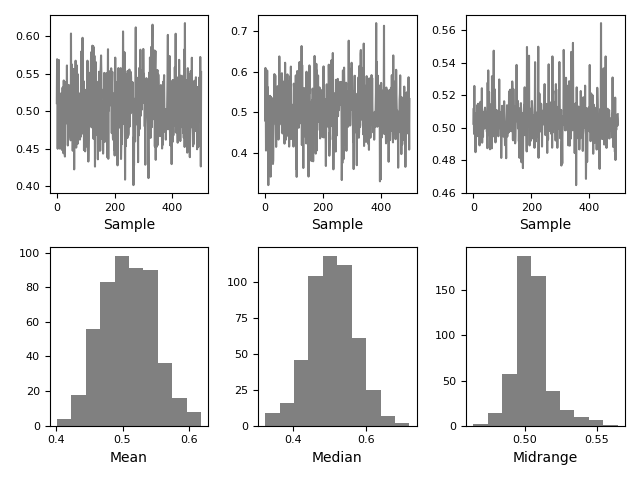
RadViz¶
RadViz is a way of visualizing multi-variate data. It is based on a simple jump tension minimization algorithm. Basically y'all set up a agglomeration of points in a airplane. In our case they are equally spaced on a unit circle. Each bespeak represents a single attribute. Yous then pretend that each sample in the information set is attached to each of these points by a spring, the stiffness of which is proportional to the numerical value of that attribute (they are normalized to unit interval). The point in the aeroplane, where our sample settles to (where the forces interim on our sample are at an equilibrium) is where a dot representing our sample will exist drawn. Depending on which class that sample belongs it will be colored differently. See the R parcel Radviz for more information.
Note: The "Iris" dataset is available hither.
In [117]: from pandas.plotting import radviz In [118]: information = pd . read_csv ( "data/iris.data" ) In [119]: plt . effigy (); In [120]: radviz ( information , "Name" ); 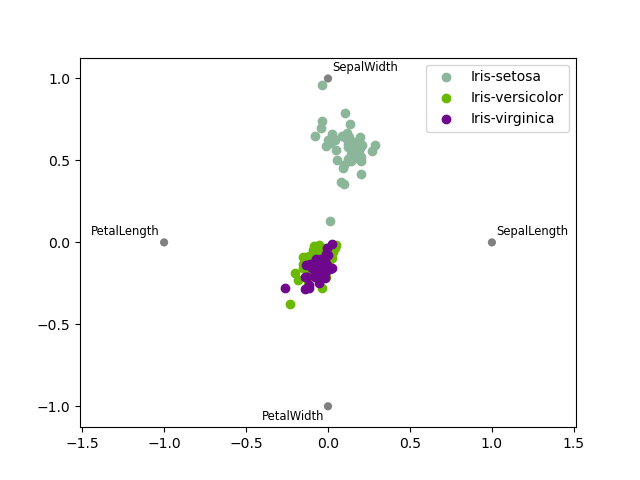
Plot formatting¶
Setting the plot style¶
From version one.5 and up, matplotlib offers a range of pre-configured plotting styles. Setting the style can exist used to hands give plots the general look that you want. Setting the mode is as easy equally calling matplotlib.style.use(my_plot_style) before creating your plot. For example you could write matplotlib.manner.utilise('ggplot') for ggplot-way plots.
You tin can see the various available style names at matplotlib.style.available and it'due south very piece of cake to endeavor them out.
General plot manner arguments¶
Nigh plotting methods have a set of keyword arguments that command the layout and formatting of the returned plot:
In [121]: plt . figure (); In [122]: ts . plot ( manner = "k--" , characterization = "Series" ); 
For each kind of plot (eastward.chiliad. line , bar , scatter ) any additional arguments keywords are passed along to the corresponding matplotlib function ( ax.plot() , ax.bar() , ax.scatter() ). These can be used to control additional styling, beyond what pandas provides.
Controlling the legend¶
You may ready the fable argument to False to hide the legend, which is shown by default.
In [123]: df = pd . DataFrame ( np . random . randn ( 1000 , four ), index = ts . index , columns = list ( "ABCD" )) In [124]: df = df . cumsum () In [125]: df . plot ( legend = Faux ); 
Controlling the labels¶
New in version 1.1.0.
Y'all may fix the xlabel and ylabel arguments to give the plot custom labels for x and y axis. Past default, pandas will pick upward alphabetize proper noun every bit xlabel, while leaving it empty for ylabel.
In [126]: df . plot (); In [127]: df . plot ( xlabel = "new 10" , ylabel = "new y" ); 
Scales¶
Y'all may pass logy to get a log-scale Y centrality.
In [128]: ts = pd . Serial ( np . random . randn ( chiliad ), alphabetize = pd . date_range ( "1/1/2000" , periods = yard )) In [129]: ts = np . exp ( ts . cumsum ()) In [130]: ts . plot ( logy = True ); 
Run across also the logx and loglog keyword arguments.
Plotting on a secondary y-axis¶
To plot data on a secondary y-axis, use the secondary_y keyword:
In [131]: df [ "A" ] . plot (); In [132]: df [ "B" ] . plot ( secondary_y = True , style = "g" ); 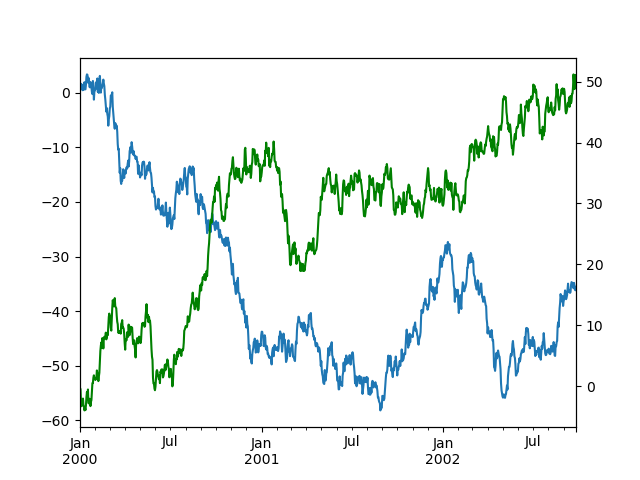
To plot some columns in a DataFrame , requite the cavalcade names to the secondary_y keyword:
In [133]: plt . figure (); In [134]: ax = df . plot ( secondary_y = [ "A" , "B" ]) In [135]: ax . set_ylabel ( "CD scale" ); In [136]: ax . right_ax . set_ylabel ( "AB scale" ); 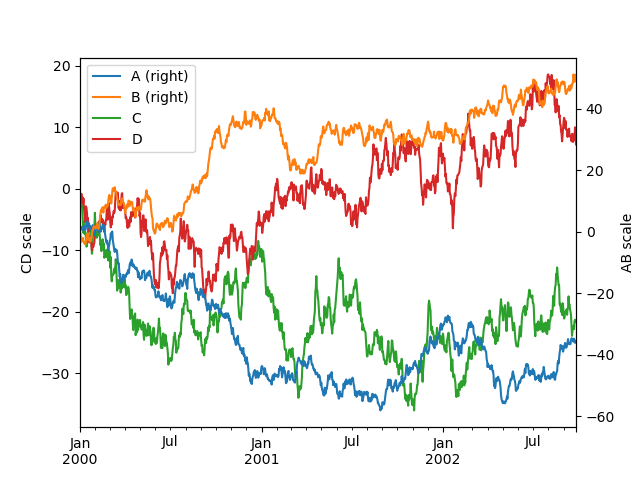
Note that the columns plotted on the secondary y-axis is automatically marked with "(right)" in the fable. To turn off the automated marking, use the mark_right=Simulated keyword:
In [137]: plt . effigy (); In [138]: df . plot ( secondary_y = [ "A" , "B" ], mark_right = Simulated ); 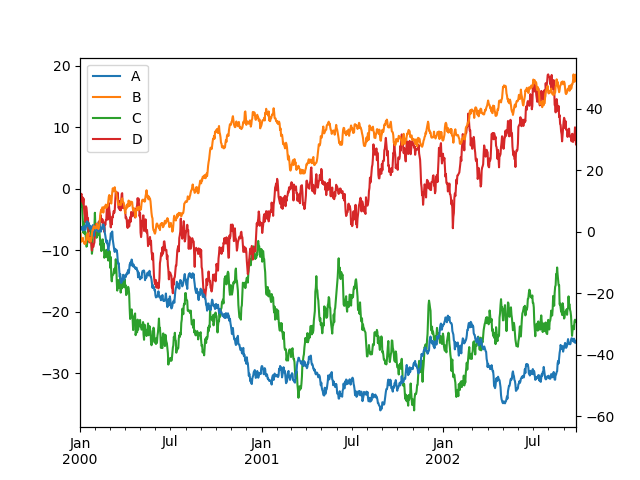
Custom formatters for timeseries plots¶
Changed in version one.0.0.
pandas provides custom formatters for timeseries plots. These change the formatting of the centrality labels for dates and times. Past default, the custom formatters are applied only to plots created by pandas with DataFrame.plot() or Serial.plot() . To take them apply to all plots, including those fabricated by matplotlib, prepare the option pd.options.plotting.matplotlib.register_converters = True or employ pandas.plotting.register_matplotlib_converters() .
Suppressing tick resolution adjustment¶
pandas includes automatic tick resolution adjustment for regular frequency time-series information. For express cases where pandas cannot infer the frequency data (e.g., in an externally created twinx ), you can choose to suppress this behavior for alignment purposes.
Hither is the default behavior, discover how the x-axis tick labeling is performed:
In [139]: plt . figure (); In [140]: df [ "A" ] . plot (); 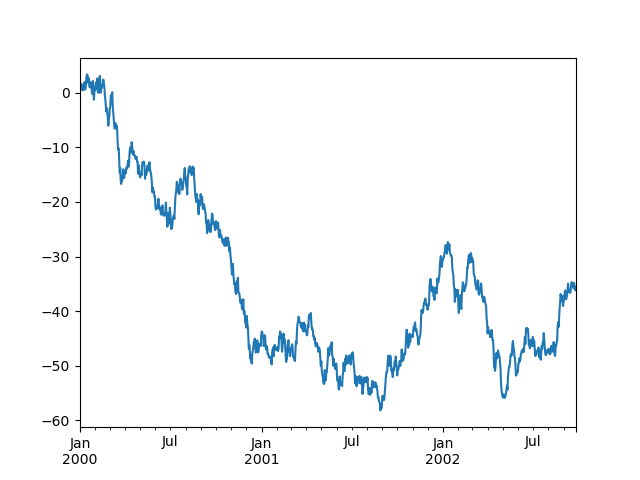
Using the x_compat parameter, you can suppress this beliefs:
In [141]: plt . figure (); In [142]: df [ "A" ] . plot ( x_compat = True ); 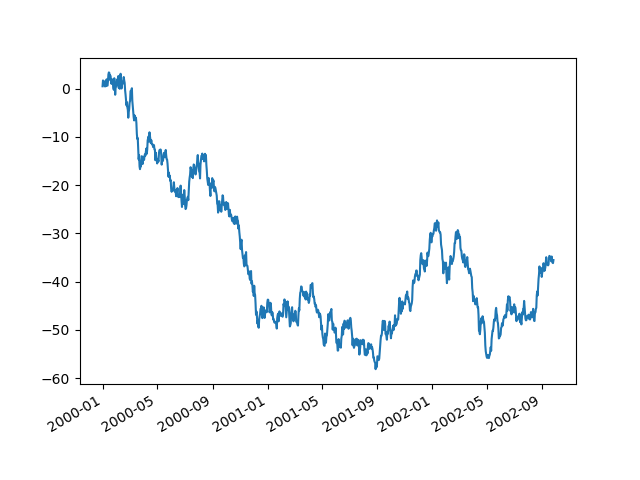
If you lot take more than one plot that needs to exist suppressed, the apply method in pandas.plotting.plot_params can exist used in a with statement:
In [143]: plt . effigy (); In [144]: with pd . plotting . plot_params . use ( "x_compat" , True ): .....: df [ "A" ] . plot ( colour = "r" ) .....: df [ "B" ] . plot ( color = "m" ) .....: df [ "C" ] . plot ( color = "b" ) .....: 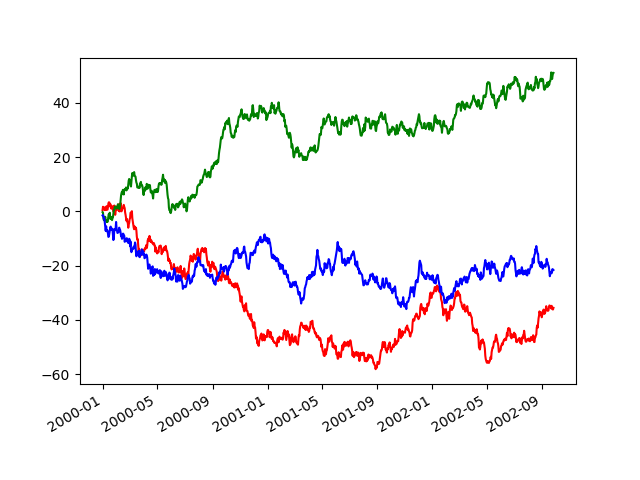
Automated appointment tick adjustment¶
TimedeltaIndex now uses the native matplotlib tick locator methods, it is useful to telephone call the automated date tick adjustment from matplotlib for figures whose ticklabels overlap.
See the autofmt_xdate method and the matplotlib documentation for more.
Subplots¶
Each Series in a DataFrame tin can exist plotted on a different axis with the subplots keyword:
In [145]: df . plot ( subplots = Truthful , figsize = ( 6 , six )); 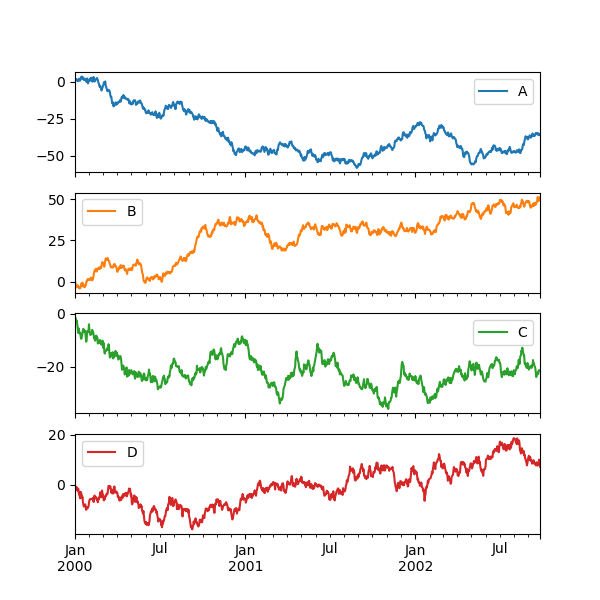
Using layout and targeting multiple axes¶
The layout of subplots can be specified by the layout keyword. It can have (rows, columns) . The layout keyword tin be used in hist and boxplot also. If the input is invalid, a ValueError will be raised.
The number of axes which can be contained past rows ten columns specified past layout must be larger than the number of required subplots. If layout can contain more than axes than required, blank axes are not drawn. Similar to a NumPy array's reshape method, you can use -1 for ane dimension to automatically calculate the number of rows or columns needed, given the other.
In [146]: df . plot ( subplots = Truthful , layout = ( 2 , 3 ), figsize = ( 6 , vi ), sharex = Imitation ); 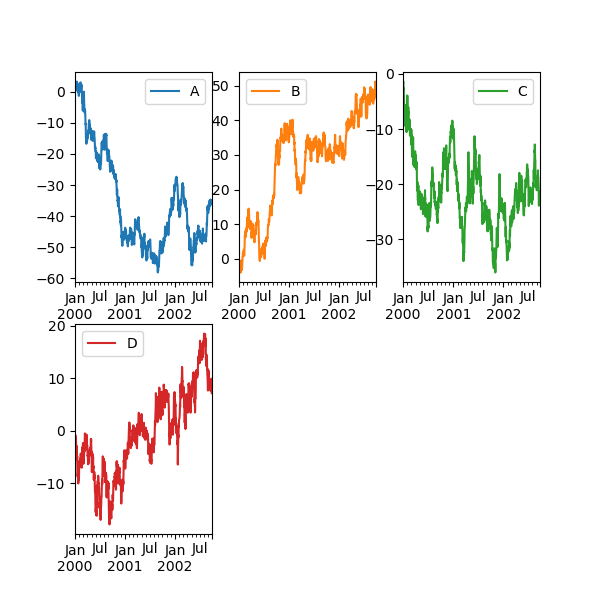
The above case is identical to using:
In [147]: df . plot ( subplots = True , layout = ( 2 , - 1 ), figsize = ( 6 , 6 ), sharex = Faux ); The required number of columns (3) is inferred from the number of serial to plot and the given number of rows (2).
You can pass multiple axes created beforehand every bit listing-similar via ax keyword. This allows more complicated layouts. The passed axes must be the same number as the subplots being drawn.
When multiple axes are passed via the ax keyword, layout , sharex and sharey keywords don't affect to the output. You should explicitly pass sharex=Faux and sharey=False , otherwise you volition see a alarm.
In [148]: fig , axes = plt . subplots ( 4 , 4 , figsize = ( 9 , 9 )) In [149]: plt . subplots_adjust ( wspace = 0.v , hspace = 0.5 ) In [150]: target1 = [ axes [ 0 ][ 0 ], axes [ one ][ 1 ], axes [ 2 ][ two ], axes [ 3 ][ 3 ]] In [151]: target2 = [ axes [ three ][ 0 ], axes [ ii ][ one ], axes [ ane ][ 2 ], axes [ 0 ][ 3 ]] In [152]: df . plot ( subplots = True , ax = target1 , fable = False , sharex = Faux , sharey = False ); In [153]: ( - df ) . plot ( subplots = True , ax = target2 , fable = False , sharex = Imitation , sharey = Imitation ); 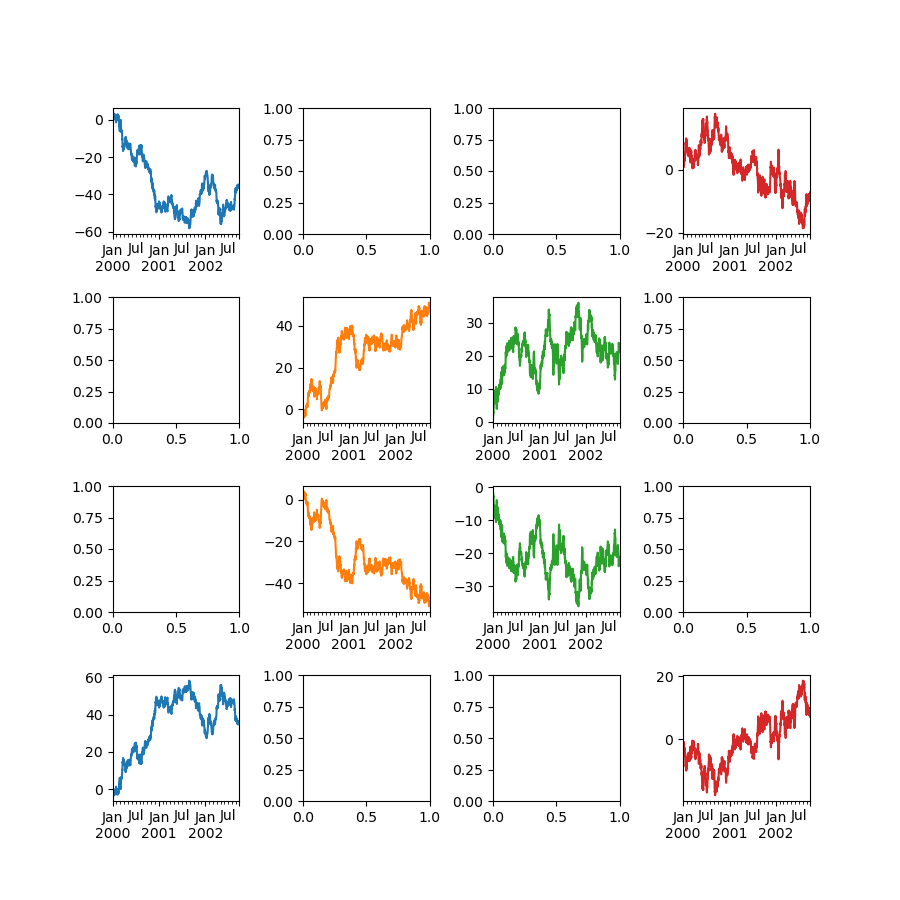
Another option is passing an ax argument to Serial.plot() to plot on a particular axis:
In [154]: fig , axes = plt . subplots ( nrows = two , ncols = 2 ) In [155]: plt . subplots_adjust ( wspace = 0.two , hspace = 0.5 ) In [156]: df [ "A" ] . plot ( ax = axes [ 0 , 0 ]); In [157]: axes [ 0 , 0 ] . set_title ( "A" ); In [158]: df [ "B" ] . plot ( ax = axes [ 0 , 1 ]); In [159]: axes [ 0 , 1 ] . set_title ( "B" ); In [160]: df [ "C" ] . plot ( ax = axes [ 1 , 0 ]); In [161]: axes [ 1 , 0 ] . set_title ( "C" ); In [162]: df [ "D" ] . plot ( ax = axes [ 1 , 1 ]); In [163]: axes [ 1 , 1 ] . set_title ( "D" ); 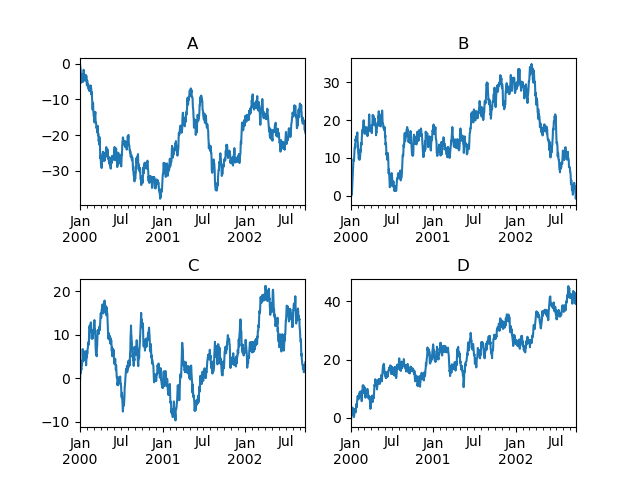
Plotting with error bars¶
Plotting with error confined is supported in DataFrame.plot() and Series.plot() .
Horizontal and vertical mistake bars can be supplied to the xerr and yerr keyword arguments to plot() . The error values tin be specified using a diverseness of formats:
-
Equally a
DataFrameordictof errors with cavalcade names matching thecolumnsattribute of the plottingDataFrameor matching theproper nameattribute of theSerial. -
Equally a
strindicating which of the columns of plottingDataFramecontain the error values. -
Every bit raw values (
list,tuple, ornp.ndarray). Must be the same length every bit the plottingDataFrame/Series.
Hither is an example of one way to easily plot group means with standard deviations from the raw data.
# Generate the data In [164]: ix3 = pd . MultiIndex . from_arrays ( .....: [ .....: [ "a" , "a" , "a" , "a" , "a" , "b" , "b" , "b" , "b" , "b" ], .....: [ "foo" , "foo" , "foo" , "bar" , "bar" , "foo" , "foo" , "bar" , "bar" , "bar" ], .....: ], .....: names = [ "letter" , "word" ], .....: ) .....: In [165]: df3 = pd . DataFrame ( .....: { .....: "data1" : [ 9 , 3 , 2 , 4 , iii , ii , 4 , half dozen , three , two ], .....: "data2" : [ nine , 6 , 5 , seven , v , four , v , 6 , 5 , i ], .....: }, .....: index = ix3 , .....: ) .....: # Group by index labels and take the means and standard deviations # for each group In [166]: gp3 = df3 . groupby ( level = ( "letter" , "word" )) In [167]: means = gp3 . mean () In [168]: errors = gp3 . std () In [169]: means Out[169]: data1 data2 letter of the alphabet word a bar 3.500000 six.000000 foo 4.666667 6.666667 b bar iii.666667 four.000000 foo iii.000000 four.500000 In [170]: errors Out[170]: data1 data2 letter of the alphabet word a bar 0.707107 1.414214 foo 3.785939 2.081666 b bar 2.081666 ii.645751 foo i.414214 0.707107 # Plot In [171]: fig , ax = plt . subplots () In [172]: means . plot . bar ( yerr = errors , ax = ax , capsize = iv , rot = 0 ); 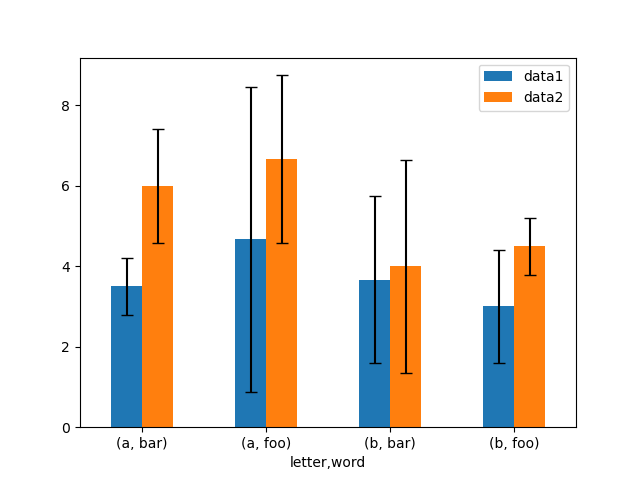
Asymmetrical error bars are also supported, however raw error values must be provided in this case. For a N length Series , a 2xN array should be provided indicating lower and upper (or left and correct) errors. For a MxN DataFrame , asymmetrical errors should exist in a Mx2xN array.
Hither is an example of one fashion to plot the min/max range using asymmetrical error bars.
In [173]: mins = gp3 . min () In [174]: maxs = gp3 . max () # errors should exist positive, and defined in the social club of lower, upper In [175]: errors = [[ ways [ c ] - mins [ c ], maxs [ c ] - means [ c ]] for c in df3 . columns ] # Plot In [176]: fig , ax = plt . subplots () In [177]: means . plot . bar ( yerr = errors , ax = ax , capsize = 4 , rot = 0 ); 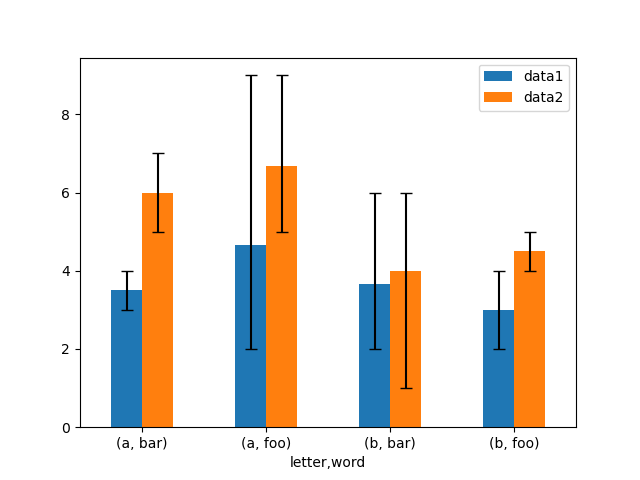
Plotting tables¶
Plotting with matplotlib table is now supported in DataFrame.plot() and Series.plot() with a table keyword. The table keyword can take bool , DataFrame or Serial . The simple style to describe a table is to specify table=True . Information will be transposed to encounter matplotlib's default layout.
In [178]: fig , ax = plt . subplots ( one , 1 , figsize = ( 7 , six.5 )) In [179]: df = pd . DataFrame ( np . random . rand ( five , 3 ), columns = [ "a" , "b" , "c" ]) In [180]: ax . xaxis . tick_top () # Brandish x-axis ticks on acme. In [181]: df . plot ( tabular array = Truthful , ax = ax ); 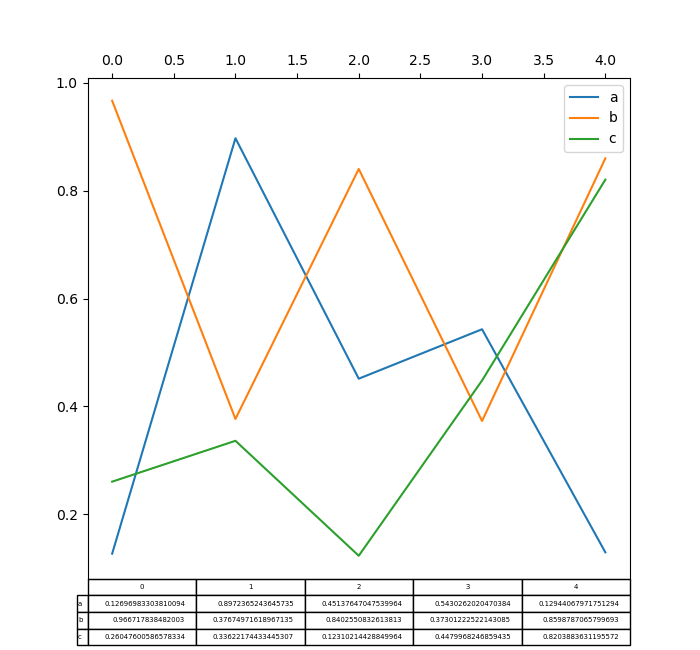
Also, you can laissez passer a different DataFrame or Series to the table keyword. The data volition be drawn as displayed in print method (not transposed automatically). If required, it should exist transposed manually as seen in the instance below.
In [182]: fig , ax = plt . subplots ( i , 1 , figsize = ( 7 , 6.75 )) In [183]: ax . xaxis . tick_top () # Display x-axis ticks on top. In [184]: df . plot ( tabular array = np . round ( df . T , two ), ax = ax ); 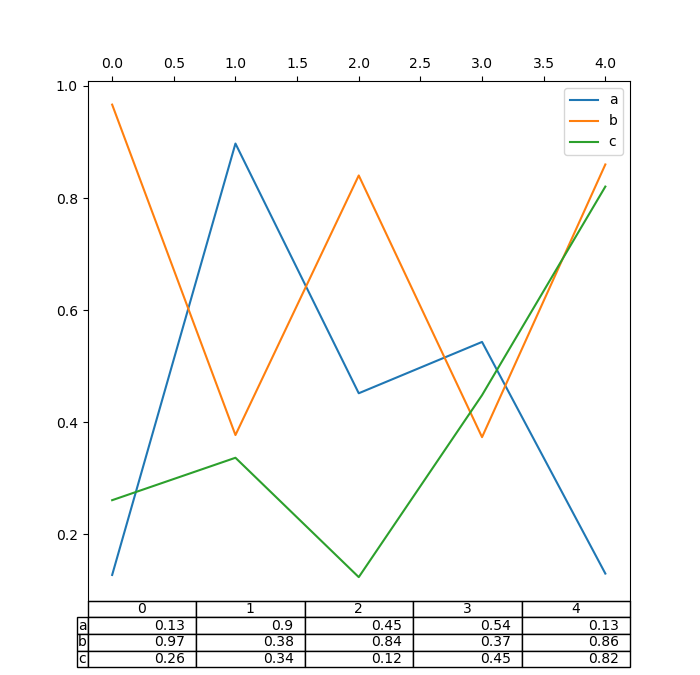
There likewise exists a helper function pandas.plotting.table , which creates a table from DataFrame or Series , and adds it to an matplotlib.Axes case. This function tin have keywords which the matplotlib table has.
In [185]: from pandas.plotting import table In [186]: fig , ax = plt . subplots ( i , 1 ) In [187]: tabular array ( ax , np . round ( df . describe (), 2 ), loc = "upper right" , colWidths = [ 0.two , 0.two , 0.two ]); In [188]: df . plot ( ax = ax , ylim = ( 0 , two ), legend = None ); 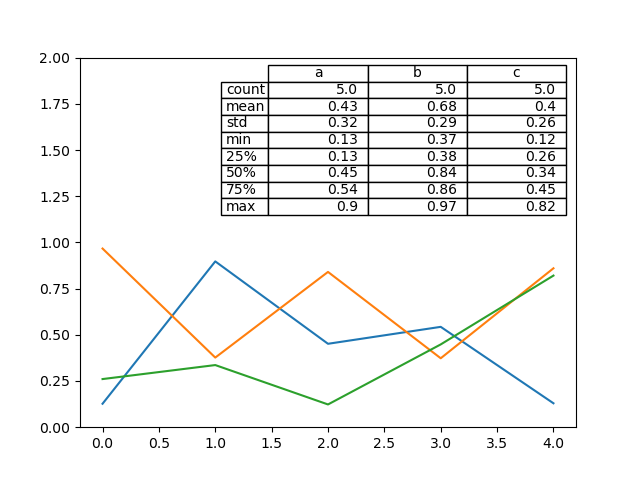
Note: You tin can get table instances on the axes using axes.tables property for further decorations. See the matplotlib table documentation for more.
Colormaps¶
A potential event when plotting a big number of columns is that it tin be difficult to distinguish some series due to repetition in the default colors. To remedy this, DataFrame plotting supports the use of the colormap argument, which accepts either a Matplotlib colormap or a string that is a name of a colormap registered with Matplotlib. A visualization of the default matplotlib colormaps is bachelor here.
As matplotlib does not directly support colormaps for line-based plots, the colors are selected based on an even spacing determined by the number of columns in the DataFrame . There is no consideration made for background colour, and so some colormaps will produce lines that are not easily visible.
To utilise the cubehelix colormap, we tin pass colormap='cubehelix' .
In [189]: df = pd . DataFrame ( np . random . randn ( thousand , 10 ), alphabetize = ts . alphabetize ) In [190]: df = df . cumsum () In [191]: plt . figure (); In [192]: df . plot ( colormap = "cubehelix" ); 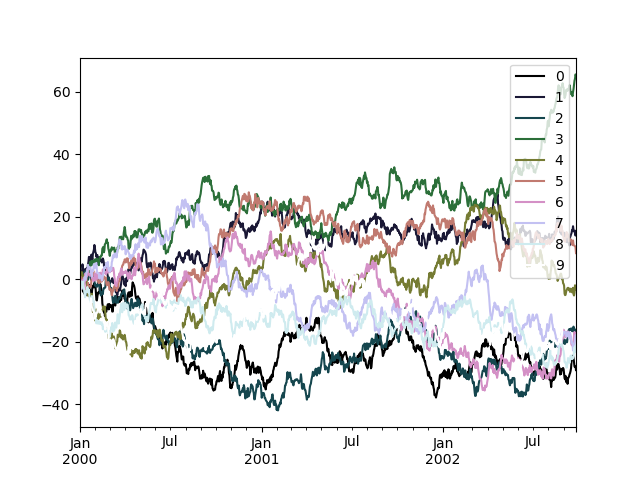
Alternatively, we can pass the colormap itself:
In [193]: from matplotlib import cm In [194]: plt . figure (); In [195]: df . plot ( colormap = cm . cubehelix ); 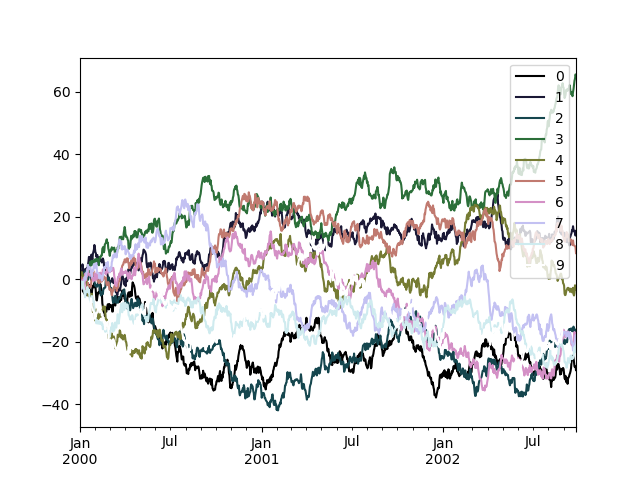
Colormaps can besides be used other plot types, similar bar charts:
In [196]: dd = pd . DataFrame ( np . random . randn ( 10 , ten )) . applymap ( abs ) In [197]: dd = dd . cumsum () In [198]: plt . effigy (); In [199]: dd . plot . bar ( colormap = "Greens" ); 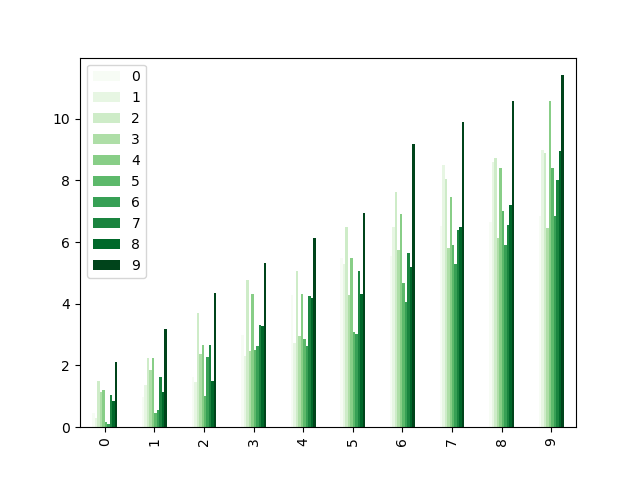
Parallel coordinates charts:
In [200]: plt . figure (); In [201]: parallel_coordinates ( data , "Name" , colormap = "gist_rainbow" ); 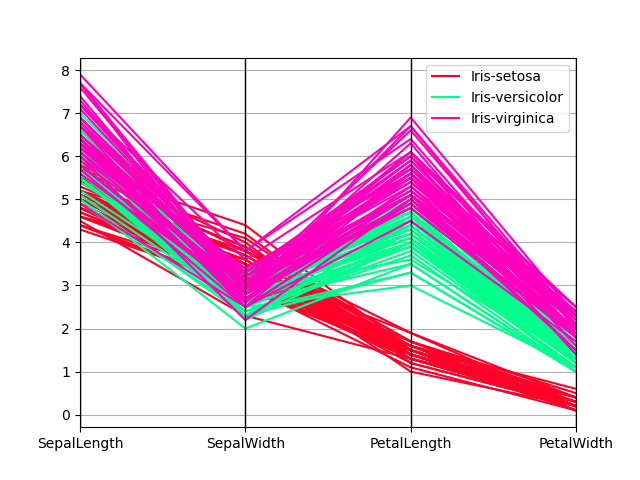
Andrews curves charts:
In [202]: plt . figure (); In [203]: andrews_curves ( data , "Proper name" , colormap = "wintertime" ); 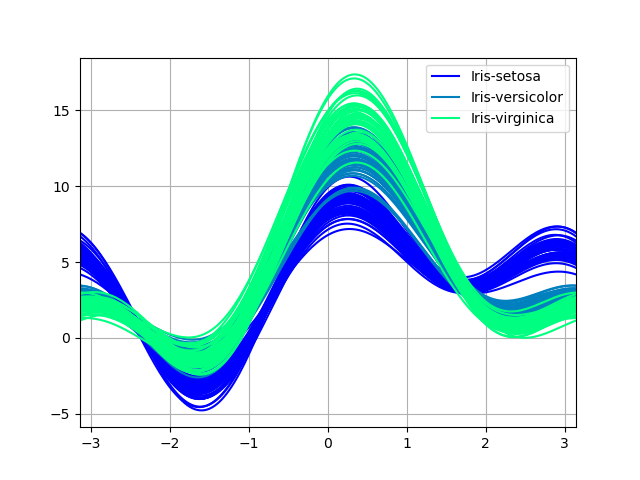
Plotting direct with matplotlib¶
In some situations it may still be preferable or necessary to prepare plots straight with matplotlib, for case when a certain type of plot or customization is not (nonetheless) supported by pandas. Series and DataFrame objects conduct like arrays and can therefore be passed direct to matplotlib functions without explicit casts.
pandas likewise automatically registers formatters and locators that recognize engagement indices, thereby extending engagement and fourth dimension support to practically all plot types available in matplotlib. Although this formatting does not provide the same level of refinement you would become when plotting via pandas, it tin exist faster when plotting a big number of points.
In [204]: price = pd . Series ( .....: np . random . randn ( 150 ) . cumsum (), .....: index = pd . date_range ( "2000-one-ane" , periods = 150 , freq = "B" ), .....: ) .....: In [205]: ma = price . rolling ( xx ) . hateful () In [206]: mstd = price . rolling ( 20 ) . std () In [207]: plt . figure (); In [208]: plt . plot ( toll . index , cost , "k" ); In [209]: plt . plot ( ma . index , ma , "b" ); In [210]: plt . fill_between ( mstd . index , ma - 2 * mstd , ma + 2 * mstd , color = "b" , blastoff = 0.two ); 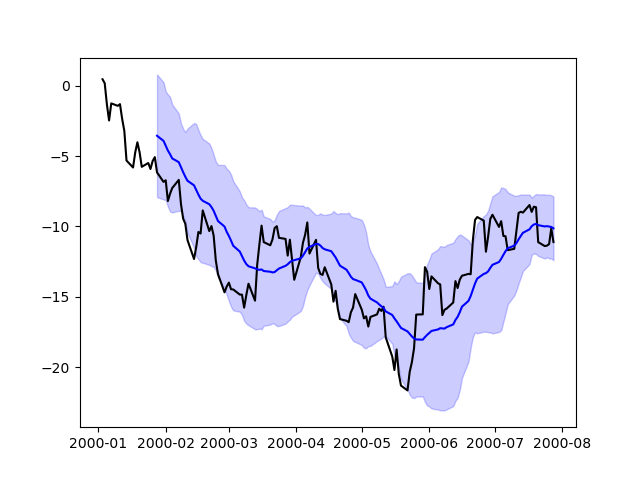
Plotting backends¶
Starting in version 0.25, pandas can exist extended with third-party plotting backends. The main thought is letting users select a plotting backend different than the provided one based on Matplotlib.
This can be done by passing 'backend.module' as the argument backend in plot function. For example:
>>> Series ([ one , 2 , 3 ]) . plot ( backend = "backend.module" ) Alternatively, y'all can also gear up this selection globally, do you don't need to specify the keyword in each plot call. For example:
>>> pd . set_option ( "plotting.backend" , "backend.module" ) >>> pd . Series ([ 1 , ii , 3 ]) . plot () Or:
>>> pd . options . plotting . backend = "backend.module" >>> pd . Serial ([ 1 , two , 3 ]) . plot () This would be more or less equivalent to:
>>> import backend.module >>> backend . module . plot ( pd . Series ([ 1 , 2 , 3 ])) The backend module can so use other visualization tools (Bokeh, Altair, hvplot,…) to generate the plots. Some libraries implementing a backend for pandas are listed on the ecosystem Visualization folio.
Developers guide can be found at https://pandas.pydata.org/docs/dev/development/extending.html#plotting-backends
Source: https://pandas.pydata.org/pandas-docs/stable/user_guide/visualization.html
0 Response to "python draw multiple histogram in 3d"
Post a Comment